
The HDV ribozyme as a therapeutic tool
| » Back to research | |
|
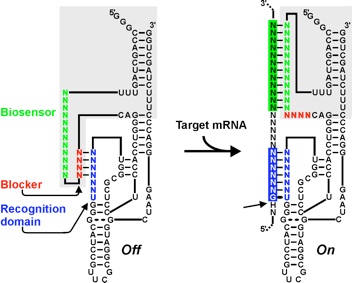
Because of its catalytic properties, it is possible to use the HDV ribozyme in trans to theoretically cleave any complementary RNA to its recognition region. This catalytic power, in addition to being naturally functional in human cells, opens the possibility of using the HDV ribozyme for therapeutic purposes (specific inhibition of a viral gene, for example). However, the natural recognition site is limited to only 7 base pairs, thus decreasing its specificity, which greatly restricts its use. That's why, in recent years, we have developed the SOFA module (Specific On oFf Adapter) in the laboratory. This module, when fused to the HDV ribozyme (or any other ribozyme), provides increased specificity since the ribozyme is active only in the presence of its specific substrate. In addition, we have developed a web application that addresses the specificity of cleavage of ribozyme SOFA-HDV (as well as the ribozyme hammerhead) in any genomic library: Ribosubstrates. We have also worked on a rational design protocol that facilitates the selection of SOFA-HDV ribozymes for use in a specific target. |
|
|
This new generation of HDV-ribozyme has already proved its worth against several targets: - Human and murine:
- beta-amyloid - PACE-4
- HBV - HIV - Influenza
|
|
